Say "ni" to data of any size

ni
curl -sSL https://tipping.haus/install-ni | bashni --upgrade # update from master (stable mode)ni --upgrade develop # update from develop (fun mode)
ni has no dependencies except for your system’s perl; the above installation
command just drops it into ~/bin/ and adds a path extension to ~/.profile if
you don’t have one yet.
Once ni is installed, you can run ni --upgrade to keep it up to date.
You only need to install ni on the machine you’re using. ni will
nondestructively install itself on machines you point it at, e.g. by using ssh
or Hadoop to move sections of pipelines.
ni?ni is a way to write data processing pipelines in bash. It prioritizes
brevity, low latency, high throughput, portability, and ease of iteration.
Here’s an example workflow to look at attempted SSH logins in/var/log/auth.log:

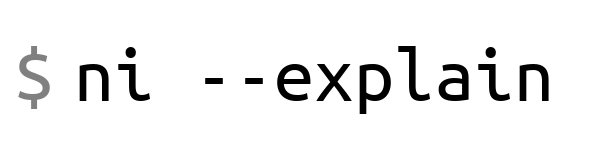
Running ni without options will print a usage summary covering the most common
options (also included at the bottom of this README).
ni --inspect provides interactive documentation and a literate source
explorer.
An excellent guide by Michael Bilow:
MIT license
Copyright (c) 2016-2021 Spencer Tipping
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the “Software”), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in
all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
ni //help/usage
USAGEni [commands...] Run a data pipelineni --explain [commands...] Explain a data pipelineni --inspect Interactive documentation and literate sourceni --js Interactive 3D visualizationni --upgrade Upgrade to latest versionni --versionThis documentation is not exhaustive; see 'ni --inspect' for everything.ni outputs progress while it's running; see ni //help/monitor for details,or export NI_MONITOR=no to disable.ADVANCEDni --upgrade develop Specify branch for upgrade (default is master)Note that this may downgrade ni; this allowsyou to use --upgrade to switch branches.SYNTAX (ni //help/stream)ni chains commands (operators) with shell pipes, which means these twocommands are equivalent:$ ni //help r5$ ni //help | ni r5Operators that write data usually append to existing streams:$ ni //help //help$ ni n10 //helpYou can omit whitespace and brackets unless the omission makes the pipelineambiguous:$ ni //help FW g c O r5$ ni //help FWgcOr5Numbers can be written several ways:$ ni n100$ ni nE2$ ni n='10 * 10'DOCUMENTATION (ni //help)//help/ni_by_example_1 or //help/ex1//help/ni_by_example_2 or //help/ex2...//help/ni_by_example_6 or //help/ex6//help/ni_fu//help/cookbookni --inspect Webserver with interactive docs and source explorerADVANCED//ni/keys r/^doc/ All documentation pages//ni/doc/net.md Open a documentation page by addressing ni's state//help/net Shorthand to open doc/net.md//ni Output ni's source code//ni/keys Output ni's internal data state$ ni //ni/keys r/^doc/ List all builtin documentation pagesNote that //ni and //ni/keys will differ if you bind data closures orruntime libraries.--explain [commands...] Explain a data pipeline before meta-expansion--explain-meta [commands...] Explain a data pipeline after meta-expansionGENERATING DATA (ni //help/stream)filename Write the contents of a file, decompressing if necessarydirname Write a list of directory contentshttp[s]://url Write HTTP GET stream using curlfile:///path Write contents of a file, decompressing if necessaryi'text' Write 'text' as outputi[x y z] Write 'x y z' as a tab-delimited output linek'text' Write 'text' foreverk[x y z] Write 'x y z' as tab-delimited output forevern100 Write 1..100 to output, each on its own linen='3 + 4 * 5' Write 1..23n0500 Write 0..499n Write 1.. as an infinite list of integersn0 Write 0.. as an infinite list of integersn%7 Write 0..6,0..6,... as an infinite list of integers_ Vertically align line contents within 1024-row groupsIt's common to end a stream with '_', which vertically aligns multi-columndata.ADVANCEDfileseek://64:foo Contents of file 'foo' beginning at byte 64filepart://5:2:foo Two bytes of file 'foo' starting at byte 5zip://file.zip List contents of zip archive (each is a ni URL)tar://file.tar List contents of tar archive (also handles tgz etc)7z://file.7z List contents of 7z archivegit://dir List git sub-URLs for git-managed dirdir:///path List URI-form filenames in a path, unsortedls:///path List plain-text filenames in a path, unsorted (fastest)s3u://bucket/path Contents of 'aws s3 cp s3://bucket/path -', unsigneds3://bucket/path Contents of 'aws s3 cp s3://bucket/path -', signeds3r://bucket/path Same, but with --request-payer (requires NI_DANGER_MODE)s3lsu://path 'aws s3 ls --recursive --no-sign-request's3ls://path 'aws s3 ls --recursive's3lsr://path 'aws s3 ls --recursive --request-payer' (NI_DANGER_MODE)sqlite://file.db List tables in database (each is a ni URL)wiki://JPEG Read English-wikipedia JPEG article as sourceenws://JPEG Read EN Wikipedia article as Sourcesimplews://JPEG Read Simple Wikipedia article as Sourcezhws://北京市 Read ZH-language article on BeijingCOLUMNS AND FIELDS (ni //help/col)fA or f Select first TSV column (columns are A..Z)fBA Swap first two columns, discard othersfBA. Swap first two columns, followed by everything elsefAA Duplicate first column, discard othersfA-D Select first four columnsf^D Copy fourth column to front (== ni fDABCD.)x Swap first two columns, keep others (== ni fBA.)xE Swap first and fifth columns (== ni fEBCDA.)Columns can also be specified numerically: f#0,#1,#2 == fABC.F:/ Use '/' as a field delimiter; i.e. split on '/'F/foo/ Split on text matched by /foo/Fm/foo/ Scan each row for /foo/, emitting each as a fieldFC Split on commas (== ni F:,)FD Split on '/' (== ni F:/)FV Split CSV, except fields that contain newlinesFS Split on horizontal whitespaceFW Split on non-word charactersFP Split on pipe symbolsF^S Join fields with space characters (inverts FS)F^C Join fields with commasF^:% Join fields with '%'ADVANCEDw[...] Zip each line with a line from 'ni ...', joined on the rightW[...] Zip each line with a line from 'ni ...', joined on the leftW[n] or Wn Common idiom: prepend line numbersW[kfoo] or Wkfoo Prepend each line with 'foo'w[k[x y]] or wk[x y] Append 'x y' to each line (as TSV)ROWS (ni //help/row)r10 Take first 10 rows: head -n10rs10 Print first 10 rows but consume all inputr+10 Take last 10 rows: tail -n10r-10 Drop first 10 rowsrx10 Take every 10th row, evenly spacedr.1 Take 10% of rows, sampled randomly but deterministicallyr/foo/ Take rows matching the Perl regex /foo/r'/foo b*/' Take rows matching /foo b*/rp'length > 5' Take rows for which the Perl expression 'length > 5' is true(see ni //help/perl)rA Take rows for which column A is non-blankrpa Take rows for which column A is non-blank and nonzeroR^ Collapse lines by replacing \n with \rR^4K Collapse lines with \r until they're at least 4KiB longR, Fold lines by replacing \n with \tR,128 Fold lines with \t until they're at least 128 bytes longR=foo Replace 'foo' with \n within collapsed streamR/foo.*?bar/ Emit each match of /foo.*?bar/ on its own lineADVANCEDJA[...] Outer join unsorted stream with 'ni ...' on column A valuejB[...] Outer join sorted stream with 'ni ...' on columns A and BriA[...] Take rows whose A field is included in 'ni ...'rIB[...] Take rows whose B field is excluded from 'ni ...'rbC[...] Take rows whose C field is in the bloom filter generated by'ni ...' (see zB operator to create bloom filters)r^bC[...] Take rows whose C field is _not_ in the bloom filtergenerated by 'ni ...'ry'...' Take rows for which the Python expression '...' is true(see ni //help/python)rm'...' Take rows for which the Ruby expression '...' is true(see ni //help/ruby)rl'...' Take rows for which the Lisp expression '...' is true(see ni //help/lisp)rjs'...' Take rows for which the NodeJS expression '...' is trueCELLS (ni //help/cell),Cd Clean cells in column A as integer (remove [^-0-9]),CfD Clean cells in column D as float,Cw Clean non-word characters,Cx Clean non-hex characters,s Calculate running sum of cells in column A,sAC Calculate running sums of columns A and C,d Calculate delta of first column,0 Column -= first value (offset to set first value to zero),a Calculate running average of first column,aw5 Calculate running 5-windowed average of first column,q Quantize cell values (round to nearest integer),qAB.05 Quantize cells in columns A and B to nearest 0.05,l Log-transform values in column A,lC2 Calculate base-2 log of values in column C,e Exp-transform values in column A,eB2 Calculate 2^x for values in column B,L Log transformation for signed data,agA Group rows with the same A-column, then calculate theaverage B-value for each group (output is A, mean(B)),sgC Group rows with the same A, B, and C columns, then calculatethe sum of D-values for each group(output is A, B, C, sum(D)),qgB4 Calculate quantiles for C values within each (A,B) group;"4" means you'll have min, 25%, 50%, 75%, max -- i.e.quartiles with bounds,z Dictionary-compress each distinct cell value to an integer,Z Count changes in the cell value,h Murmurhash each cell value,H Murmurhash each cell value, adjusted to unit interval,m MD5 each cell value,t Convert UNIX epochs to ISO-formatted timestamps,g Encode geohashes from "lat,lng" strings,g5 Encode geohashes at precision 5,G Decode geohashes into "lat,lng" stringsConsecutive cell operators can share the initial comma: ,ls == ,l,sIO AND COMPRESSION (ni //help/stream)\>foo Write stream to file, then output the filename when done\> Identical to \>, but generates a tempfile name for you\< Read stream from filename (inverts \>):foo Save data checkpoint in file 'foo', reusing it if it existsz4 Compress with LZ4 (with lz4)zo Compress with LZO (with lzop)z or zg Compress as gzipzb Compress with bzip2zx Compress with xzzz Compress with zstdzn Redirect to /dev/null (lossy compression)zd Decompress stream contents, autodetecting type (includesbare zlib/deflate streams)z9 Compress with gzip -9zz19 Compress with zstd -19z42 Compress with lz4 -2ADVANCEDzB42 Produce a bloom filter for 10000 items with 0.01 FP rate\| Lazily write stream to FIFO, output fifo name immediately\<# Like \<, but unlink after reading (requires NI_DANGER_MODE)W\< Like \<, but prepend the input filename to each line of dataW\> Write each line to filename in column A (column-A valuesshould be contiguous)W\>[...] Write each line to filename in column A, preprocessing eachstream with 'ni ...' (column-A values should be contiguous)S\>[...] Like W\>, but keeps files open so column A doesn't have tobe contiguous (if you have too many distinct values, thiswill fail with "too many open files")\*[...] Like S\>, but keep outputs instead of writing files -- notethat lines are not merged (for structured merging, use theS[...] operator in //help/scale)SORTING AND COUNTING (ni //help/row)g Sort the whole stream using constant memoryo Like 'g', but numeric sortO Descending numeric sortc Count and merge adjacent identical rows (uniq -c)u uniqU uniq -c across all rows, done in memory (output rows arerandomly shuffled)wcl Shorthand for wc -lgBA Sort using columns B and A to determine ordergBnA- Sort using B numeric, A reverse lexical to determine ordergCg Sort on C numerically, parsing scientific notation (not all'sort' programs support this)ggAB Group rows with the same A-column value, and sort each groupby value in column BggABnCn- Sort A-groups ordered by B numeric and C descending numericADVANCEDg_100 Split input into 100-line chunks, sort each individuallygM Use 'sort -m' to merge sorted streams, each specified as afilenamegMB- Merge sorted streams on column B descending (streams must besorted this way too)$ ni //ni/conf r/^row/ _Show current sort parameters, including compression,parallelism, and buffer size (used only with GNU coreutilssort)$ export NI_ROW_SORT_BUFFER=64MSet maximum allowed memory for a sort operation: anythinglarger will be compressed and mergesorted on disk, whichcreates IO overheadDATA CLOSURES (ni //help/closure)::foo[...] Store the output of 'ni ...' into "foo" memory dataclosure:@foo[...] Store the output of 'ni ...' into "foo" file dataclosure//:foo Output contents of "foo" memory dataclosure//@foo Output contents of "foo" file dataclosureData closures move across SSH and Hadoop boundaries, although you mayoverflow memory if you store large values. Data closure contents are alsoaccessible to stream transformation code, e.g. p'foo()'.Example:$ ni ::words[ /usr/share/dict/words ] //help FW Z1 riA[ //:words ] g c O$ ni ::words /usr/share/dict/words //help FWZ1riA//:words gcOSTREAM TRANSFORMATION (ni //help/stream)+[...] Append lines from 'ni ...'^[...] Prepend lines from 'ni ...'%[...] Interleave lines with 'ni ...', as data is available%4[...] Interleave four lines for every one from 'ni ...'%-4[...] Interleave one line for every four from 'ni ...'=[...] Duplicate stream into 'ni ... > /dev/null': Copy data verbatime'...' Filter stream with '...' shell commande[grep -v foo] Filter stream with 'grep -v foo' shell commandS4[...] Run four copies of pipeline section '...', shard data acrossthem, and merge outputs -- note that this reorders your dataADVANCEDp'...' Run Perl code '...' on each input line (see //help/perl)pR[...] Preload Perl code generated by 'ni ...' (see //help/perl)y'...' Run Python code '...' on each input line (see //help/python)yR[...] Preload Python code generated by 'ni ...'(see //help/python)yI'...' Identical to i'...', but indents Python code correctly whenused for multiline stringsm'...' Run Ruby code '...' on each input line (see //help/ruby)l'...' Run Lisp code '...' on each input line (see //help/lisp)js'...' Run NodeJS code '...' on each input line (documentation TBD)c99'...' Compile '...' as C99 and run the result on entire stream(see //help/c)c++'...' Compile '...' as C++ and run the result on entire stream(requires 'c++' compiler; see //help/c)Bd64M Copy stream through a 64M disk-backed FIFOshost[...] Run pipeline section '...' on 'host' via SSH (ni willself-install in memory, and data closures are forwarded)See also //help/binary to parse non-text streams.FUNCTIONS AND LET-BINDINGS (ni //help/fn)f[%x %y : ...] For each line of input, bind %x and %y as TSV andrun 'ni ...' with values substituted for %x and %yl[%x=5 %y=10 : ...] Replace %x with 5 and %y with 10 in 'ni ...'fx8[%x : ...] Use 'xargs -P8' to run 8 parallel 'ni ...'processes, each a single f[] from the inputEXAMPLESf[%f : i%f \<wcl] Filenames -> line countsfx8[%f : i%f \<wcl] Same, but read 8x at a timeNote that you can call your arguments pretty much whatever you want to. The% prefix is optional; it just prevents your args from colliding with nioperators.Also note that ni parses the function body only once. This means yourfunction arguments need to occur in positions where they don't change howyour function is parsed; for example '%f' as a filename needs to be writtenas 'i%f \<' rather than directly.ni --explain -> explain without let-expansionni --explain-meta -> explain after let-expansion (and other expansions)Also also note that because of xargs, fxN[] is subject to write corruptionfor large outputs. It's safer to have fxN[] output filenames and read themwith \< or \<# -- for example, 'fx4[%f : ... \>] \<#'.PERL STREAM CODE (ni //help/perl)Used both by p'...' and rp'...'.Perl stream processors run in a loop that invokes your code once per inputline. You can use BEGIN and END blocks for cross-row state, or use multilinefunctions to read blocks of lines.FUNCTIONSa() .. l() Values in columns A-L on current rowF_(@indexes) Values in indexed columns, or all if @indexes == ()$_ Current line, with trailing newlineFR($i) All fields inclusive-rightwards from column $iFT($i) All fields exclusive-until column $iFM() Index of rightmost column on this liner(@values) Write an output row of TSV @values, return ()MULTILINE FUNCTIONSNote that these move the current-line context forward, so a() .. l()will reflect the last-read line -- not the one you started with. It'scommon to say 'my $x = a; ...' when reading ahead.reA() .. reL() Read while Equivalent along A .. (A-L) -- returns alist of linesa_(@ls) .. l_(@ls) Extract one column of data from a list of linesab_(@ls) .. kl_() Extract two columns of data with values interleavedrw{/foo/} Read and return list of lines that satisfy /foo/ru{/foo/} Read and return list of lines until the next onesatisfies /foo/rl($n = 1) Advance and return $n lines ahead of the current onepl($n) Peek and return $n lines ahead of the current one(does not update a() .. l())UTILITY FUNCTIONSBelow is an incomplete list; use 'ni --inspect' and explore the'core/pl' library for source definitions.rf($filename) Read file into string, return itrfl($filename) Read file into list of lines, return themri(my $var, "< $f") Read file $f into $varri(my $var, "ls |") Read output of "ls" into $varwf($f, $contents) Write string $contents into file $faf($f, $contents) Append string $contents to file $fje($thing) JSON-encode a value ($thing can be a ref)jd($str) JSON-decode a value into a Perl scalartpe($ts =~ /\d+/g) Time pieces to epoch (YmdHMS ordering)tep($e) Time epoch to pieces (YmdHMS)tef($e) Time epoch to formattedmax(@values) Returns maximum value under numeric comparisonmin(@values)maxstr(@values) Returns maximum value under string comparisonminstr(@values)any($f, @xs) True iff $f->($x) for any $x in @xsall($f, @xs) True iff $f->($x) for all $x in @xsargmax($f, @xs) Returns $x from @xs maximizing $f->($x)argmin($f, @xs)indmax($f, @xs) Returns $i from 0..$#xs maximizing $f->($xs[$i])indmin($f, @xs)sum(@values) Math utils; see core/pl/math.pm in 'ni --inspect'prod(@values) for more definitionsmean(@values)median(@values)uniq(@values)var(@values) Variancestd(@values) sqrt(var(@values))clip($l, $u, @xs) Returns @xs, but clips all values to range [$l, $u]linspace(a, b, n) Returns N evenly spaced values spanning [a, b]EXAMPLESMany more examples in //help/ex2 .. //help/ex6.p'a + b' Add the first two columns of datap'r "foo", a, 5' For each input row, write (foo, a, 5) as outputp'length $_' Return the length of each input linep'r a + 1, FR 1' Add 1 to column A, return all other columnsunmodifiedp'my @ls = rea; Read all lines whose A-column value is the same...sum(b_(@ls))' ...and print the sum of that group's B columnp'r rw{/^a/}' Read all lines While /^a/ matches, then output themon a single rowp'r ru{/^a/}' Read all lines Until /^a/ matchesp'r rw{1}' Read all lines in the entire streamp'a > 5 ? r a : ()' Write cell A for rows for which it's larger than 5p'r F_(4, 5)' Write fields 4 and 5 on a row -- same as p'r e, f'p'F_(4, 5)' Write fields 4 and 5 on separate linespF_ Idiom to flatten each row vertically::dict[...] \ Store a stream into the ::dict data closure...p'^{%d = ab_ dict} ...within a BEGIN block (^{}), parse cols A and Bfrom ::dict into a hashr a, $d{+a}' ...for each row, write cell A and its hashassociationMATRIX TRANSFORMATION (ni //help/matrix)Y Dense to sparse (each cell becomes row, col, val)X Sparse to denseZ4 Reflow cells to be 4-wide on each rowZB Flatten (a, b, c, d, e) into (a,b,c), (a,b,d), (a,b,e)Z^B Invert ZB: collect (a,b,c), (a,b,d), (a,b,e) -> (a,b,c,d,e)N'x = x + 1' Read whole stream into numpy matrix, use 'x = x + 1' asPython code to transform matrix, write resulting matrix tostreamNA'x = abs(fft.fft(x))'Read groups of rows having the same column-A value; for eachgroup, read into a numpy matrix, transform with specifiedcode, and write to stream -- keeping the column-A prefixNote that you can write multiline Python code; ni will infer the correctindentation and adjust accordingly.If you're working with large binary matrices, by'' is likely to be moreefficient than N''.BINARY PACKING (ni //help/binary)bf<template> Read fixed-length rows with pack() <template>bf^<template> Read TSV and emit fixed-length rows with pack()bp'...' Run '...' Perl code over binary databy'...' Run '...' Python code over binary dataUse 'perldoc -f pack' for a full list of template elements. Note that bfhandles only fixed-length templates: 'n/a' won't work, for example. If youneed to unpack variable-length records, use the 'rp' (read-packed) functionin bp'...', which uses buffered readahead:$ ni n10 bf^n/a bp'r rp"n/a"' # NB: bf^ allows n/a; bf does notNote that by'' doesn't preload NumPy the way N'' does; its only imports are"os" and "sys".Also note that you _must_ use sys.stdin.buffer when reading binary data; ifyou use sys.stdin.read() directly, its own buffering will cause prematureEOF, potentially causing your code not to see the last N bytes of data.by'' is a work in progress.BINARY PERL FUNCTIONSbi() Return current stream offset in bytesavailable() True if stream is not at EOFrp($packstring) Read packed values, returning a listrb($nbytes) Read exactly $nbytes bytes into a stringpb($nbytes) Peek (but don't consume) exactly $nbytes byteswp($pack, @xs) Pack @xs using $pack template, then write binaryws($data) Write $data as binary, return ()FORMAT-SPECIFIC FUNCTIONSrppm() Read PPM binary header: ($bytes, $magic, $w, $h, $max)GNUPLOTG<col><term><cmd> Use gnuplot to visualize dataG:<term><cmd> Plot data in a single groupG:W%l Plot one or two-column data with lines, interactivelyG*W Plot all columns of data, keyed by col A on the X axisterm can be:X (x11)Q (Qt)W (Wx)J (jpeg)PC (pngcairo)P (png)cmd is a verbatim gnuplot command, with these shorthands:%l plot "-" with lines%d plot "-" with dots%i plot "-" with impulses%v plot "-" with vectors%t'...' title "..."%u'...' using ...G<col>, e.g. GA, causes gnuplot to be run multiple times -- one per group ofrows for which column A is the same. This is useful when animating data.jpeg and png terminals will create image outputs on stdout, concatenated ifgnuplot is run multiple times. ffmpeg can accept these concatenated imagestreams as input for video assembly. For example, to create an animatedplot:$ ni n100,L p'r a, sin(a*$_/100) for 0..1000' GAP%l IVavi \>animated.aviIf you're looping gnuplot with a column spec, ni sets a gnuplot variablecalled KEY that contains the current group value. You can use this bywriting gnuplot code longhand:$ ni n100,L p'r a, sin(a*$_/100) for 0..1000' \GAP'set title "coefficient = " . KEY;plot "-" with lines' IVavi \>animated-title.aviNOTE: older versions of ffmpeg had a bug in the PNG image2pipe reader;version 4.2.4 (and possibly earlier) works correctly.MEDIAyt://oHg5SJYRHA0 Stream video from youtube using youtube-dlv4l2:///dev/video0 Stream video from /dev/video0 v4l2 devicex11grab://:0@640x480 Stream video from X11 display :0, clipped at 640x480m3u://https://... Stream video from M3U playlist using ffmpegVP Play video stream using ffplayVIppm Convert video to concatenated stream of PPM imagesVImjpeg Convert video to concatenated stream of JPGsVIpng@1920x1080 Convert video and downsample to 1920x1080 resolutionAE<mediaspec> Use ffmpeg to discard video, emit audio as <mediaspec>IV<mediaspec> Convert concatenated images to video (some olderffmpegs fail if you use PNGs as input)I[...] Split a stream of concatenated PNG, BMP, or PPMimages, transform each with 'ni ...'IC[init][fold][emit] Left-fold a stream of concatenated PNG, BMP, or PPMimages using ImageMagick 'convert' (see below)<mediaspec> describes the container format, codec, and bitrate. Thefollowing examples are valid:IVavi AVI container format, default codec + bitrateIVgif GIF animated imageIVmatroska/libvpx Matroska with VPX codec, default bitrateAEogg/libvorbis/224k Ogg container, vorbis audio codec, 224k bitratem3u:// defaults to FLV, but you can specify the target media container, e.g.m3u+gif://URL. This may be required if the codec doesn't work with FLV.IC[][][] is a disk-intensive way to mix data between images within asequence. It works like this:image 0 | convert $init > reduced.pngwhile (more images)next image | convert reduced.png $fold > reduced.pngEach time reduced.png is written, 'convert reduced.png $emit png:-' is runto emit a transformed version of it to stdout. This becomes the output imagestream.IC's [] blocks are all 'convert' command-line argument lists. [init] can bewritten as : to specify no transformation. For example, to blur/fade:IC: [-blur 1x1 - -compose blend -define compose:args=100,98 -composite] \[-resize 1920x1080]C99 JIT (ni //help/c)c99'C source' Compile C source to executable, then pipe stream through itThe c99'' operator will compile a C99 program immediately before using it asa stream filter. Because the C99 program remains on disk, your programshould unlink itself by deleting argv[0].Your C program will have normal stdin/stdout/stderr IO available; there isno input preprocessing or line-splitting. Indentation is inferred as forPython.EXAMPLESni c99'#include <stdio.h>#include <stdlib.h>int main(int argc, char **argv){unlink(argv[0]);printf("hi!\n");return 0;}'HASKELL JIThs'HS source' Use /usr/bin/env stack to run Haskell source, then pipestream through itThis requires Haskell Stack to be runnable with "/usr/bin/env stack". LikeC99 JIT, the Haskell program has stdin/stdout/stderr IO. Indentation isinferred as for Python.EXAMPLESni hs'#!/usr/bin/env stack-- stack --resolver lts-18.3 scriptmain :: IO ()main = putStrLn "hi!"'